Advance/NanoLabo is a software for first-principles calculations and molecular dynamics, “designed for beginners.”
Advance/NanoLabo is a Graphical User Interface (GUI) designed to work with open-source materials analysis software, such as Quantum ESPRESSO and LAMMPS. You can search material databases such as Materials Project and
easily set up modeling and computational conditions. First-principles
and molecular dynamics calculations can be performed, and the results
are visualized instantly. The
intuitive and user-friendly GUI, designed for beginners, has garnered
support from many users, with hundreds of sales both domestically and
internationally since its launch in 2018.
Features of Advance/NanoLabo
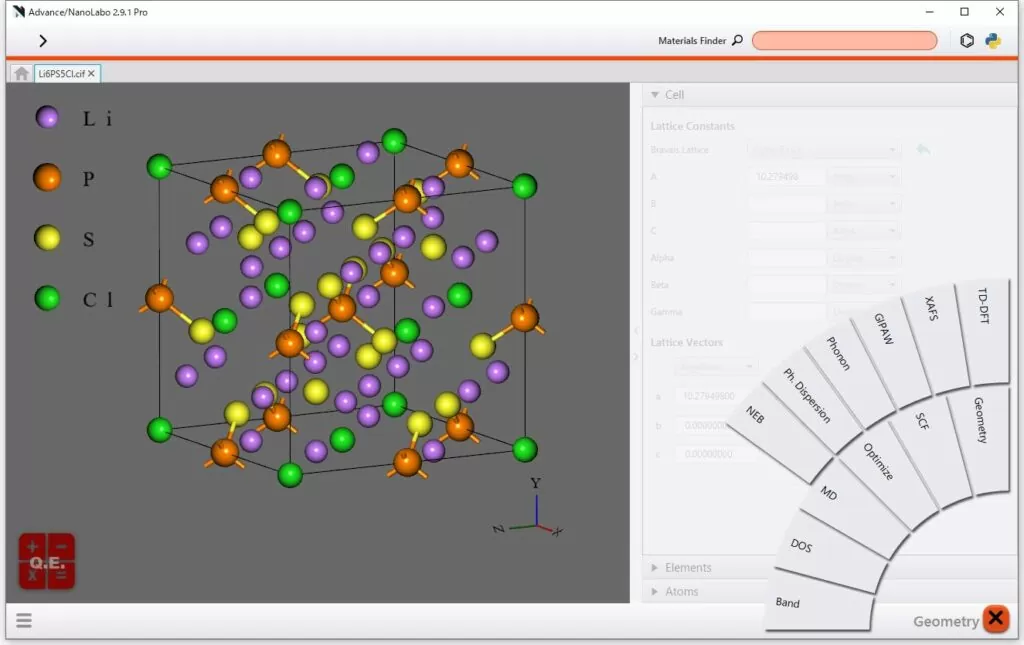
1. Intuitive and User-Friendly GUI
It is designed to be intuitive and requires only a minimum number of
steps, making it ideal for those new to material simulation. See more →
|
 |
|
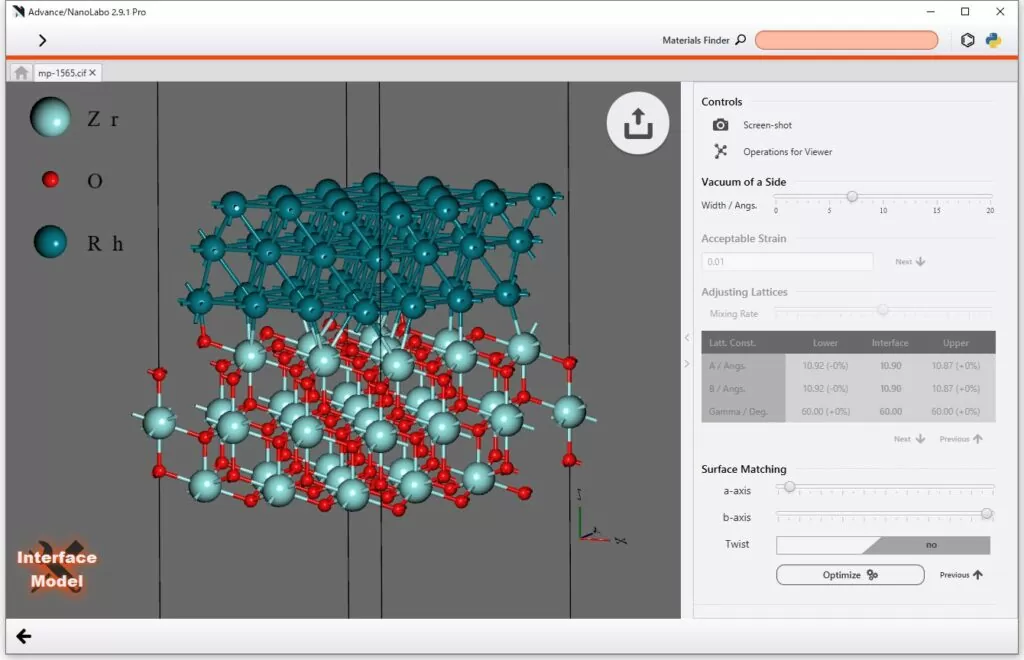
2. Versatile Modeling Capabilities
Supercells,
impurity substitution, lattice defects, space group determination,
surface and interface models, organic molecule drawing, molecular
adsorption, solvent filling, polymer modeling, and more. See more → |
3. Compatible with NN Force Fields
In addition to open-source general Graph Neural Network force fields(Open Catalyst, M3GNet, CHGNet), you can use force fields created with our proprietary Advance/NeuralMD. The force field database is also available publicly.
|
|
The majority of users are experimental researchers.
Many
software for first-principles calculations and molecular dynamics are
designed for theoretical and computational researchers, and are not
suitable for experimental researchers to conduct material simulations
alongside their primary work. However, our Advance/NanoLabo is designed
as a truly user-friendly GUI that can be easily handled even by beginners, such as experimental researchers. In fact, as of December 2023, 54% of our several hundred users are experimental researchers.
Details of AdvanceNeuralMD
|
Technical Specifications
Execution of Calculations
|
| Calculation Engine |
Quantum ESPRESSO (First-Principles)
LAMMPS (Molecular Dynamics)
ThreeBodyTB (Genaral-Purpose Tight-Binding Method) |
| Calculation Functions |
SCF Calculation, Structural Optimization, Hybrid Functional、vdW Correction,
Band Structure, Density of States (w/ PDOS Calculator), Visualization of Charge Density,
First-Principles MD, Car-Parrinello MD, Classical MD, MD with Neural Network Force Fields,
Thermal Conductivity, Viscosity Coefficient, Diffusion Coefficient, Radial Distribution Function,
TD-DFT, XAFS/EELS, Phonon (Effective Charge, Dielectric Constant, Band Structure, Density of States),
NEB Method, Work Function (ESM Method) |
| Resources |
Local Machine
Calculation Server (Supports SSH Connection and Job Management with PBS/SLURM/PJM)
Cloud
(Mat3ra, Science Cloud GPU)
NanoLabo Cloud Desktop |
modeling
|
| Materials Database |
Materials Project
PubChem |
| Crystal Systems |
Cell Translation, Supercell, Impurity Substitution,
Lattice Defects, Space Group Determination, Primitive Cell
Transformation, Standard Cell Transformation |
| Surface and Interface Systems |
Surfaces with Any Orientation, Molecular Adsorption on Surfaces, Mismatched Interfaces (Pro Version Only) |
| Molecular Systems |
Drawing Organic Molecules, Filling Solvent Molecules, Polymer Models (Pro Version Only) |
| System Requirements |
| OS |
Windows 10/11 (64bit)
AlmaLinux 8 (64bit)
macOS 13 or higher (Intel/ARM64) |
Machine Spec
(Recommended) |
CPU : Intel Core i7 or higher
Memory : 10 GB or more |
| Click here
to know more this product. |
|
Jupyter Interface for NanoLabo
This is a service that allows you to display and operate Jupyter Lab on
the screen of Advance/NanoLabo. It transfers the structure files
modeled in Advance/NanoLabo to the server where Jupyter Lab is
operating and outputs Python code that generates ASE‘s Atoms objects.
It is also possible to integrate with the general-purpose atomic-level simulation cloud service Matlantis, provided by Preferred Computational Chemistry Corporation.
|
| Click here
to know more about Jupyter Interface. |
|


