
The Computational Chemistry toolkit.
From HPC clusters to the world in one click.
Create module walkthrough
|
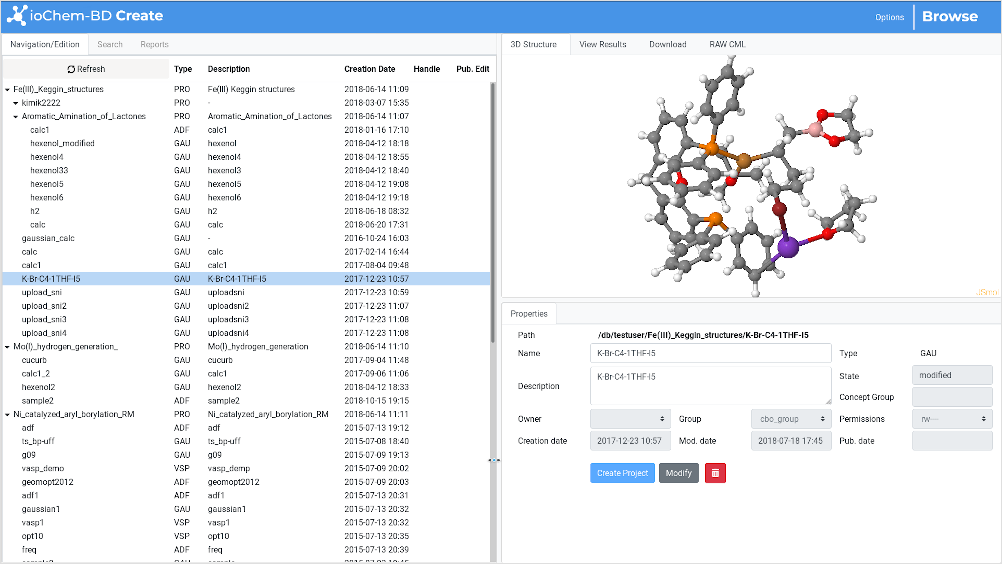 |
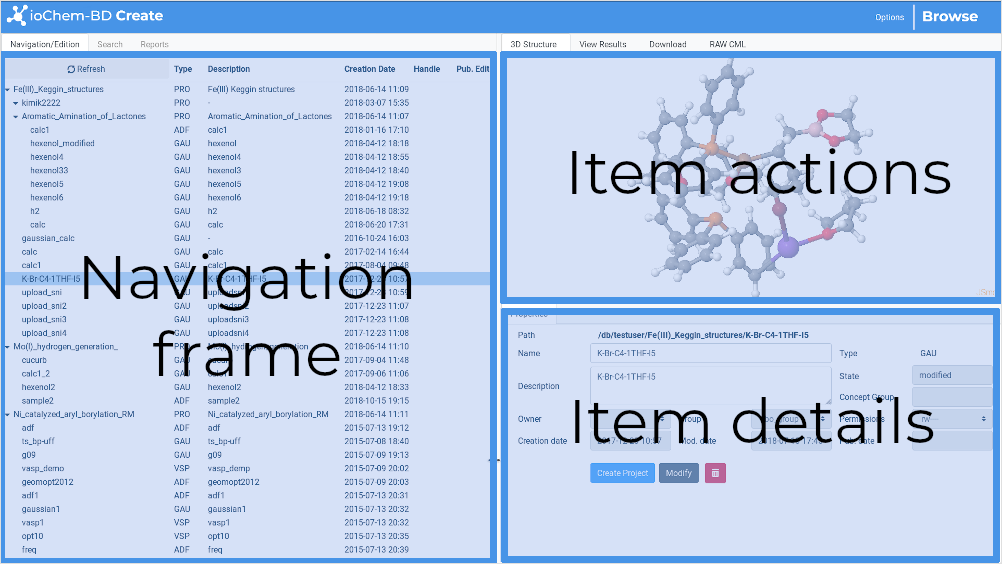
The window is composed of three frames, some with several tabs:
Navigation / Search / Reports frame
Item Actions frame
Item Details frame
The default operation flow within the Create module
starts by navigating/browsing user-uploaded content in the Navigation
frame and selecting one of them to display its content.
When any Item is selected, both the Item Actions frame and the Item Details frame get actualized and display additional data.
Multiple element selection is allowed, this is normally associated to
global operations, such for instance when publishing a project, or when
generating reports. Multiple element selection also disables Item
Actions and Item Details frames.
|
|
Navigation frame
This frame is the entry point of all managing functionalities of the Create module.
Three tabs allow us to select the current operative mode:
To switch between modes, the user only has to click on each tab: |
|
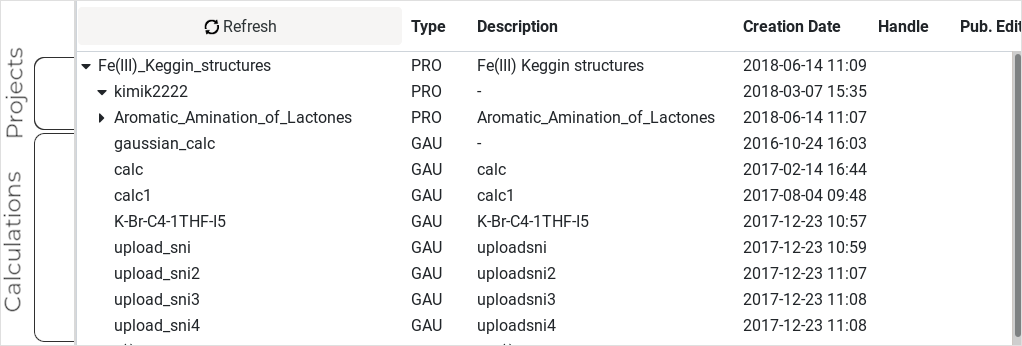
Each node represents what we call a project. This is
just a container element, like a folder, and can contain other
subprojects or simply a set of single calculations, which are the basic
element of your data.
A calculation usually contains input files, CML converted output files, additional files, etc ...
The Navigation frame is the place to gather projects and calculations
and organize your data sets in the way you want to eventually publish
them.
We can easily recognize projects inside the workspace by looking at the
Type column as they are labeled as PRO. Another way to recognize them
(when they contain child elements) is by a small arrow next to their
name. When clicking on it, the project's content will expand or
collapse.
Calculations are the leafs in the hierarchy tree, ending points where
nothing can hang from them. A calculation item represents all data
associated to a single calculation, which comes straight from the
output of computational chemistry packages. By selecting a calculation,
both the Item Actions frame and the Item Details frame get actualized
to display additional data.
On calculations, the Type column contains the abbreviation of the original software package used:
- ADF/AMS: Calculation generated with Amsterdam Density functional software package
- AMB: Calculation generated with Amber software package
- CAS: Calculation generated with CASTEP software package
- GAU: Calculation generated with Gaussian software package
- GMC: Calculation generated with GROMACS software package
- GRO: Calculation generated with GronOR software package
- GRRM: Calculation generated with GRRM software package
- LAM: Calculation generated with LAMMPS software package
- MOL: Calculation generated with Molcas software package
- MOP: Calculation generated with Mopac software package
- ORC: Calculation generated with Orca software package
- QEX: Calculation generated with QuantumESPRESSO software package
- SIE: Calculation generated with SIESTA software package
- TML: Calculation generated with Turbomole software package
- VSP: Calculation generated with Vienna Ab initio Simulation Package software package
This list will increase as more calculations types are implemented.
Other interesting columns that are worth explaning are:
|
 |
- Description: Text about the project/calculation (special symbols are allowed).
- Status: Current element status. Possible values: created/published.
Published flag: Check mark indicating if the element is published
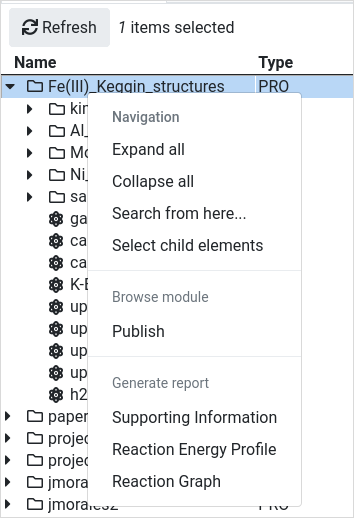
There
is a context menu to help you with the repetitive actions. You have to
first select any element in the workspace with the left click and then
right click on it to see its available options in the context menu.
Read the Publishing Calculations section or the Generate reports section to learn how to proceed further. |
 |
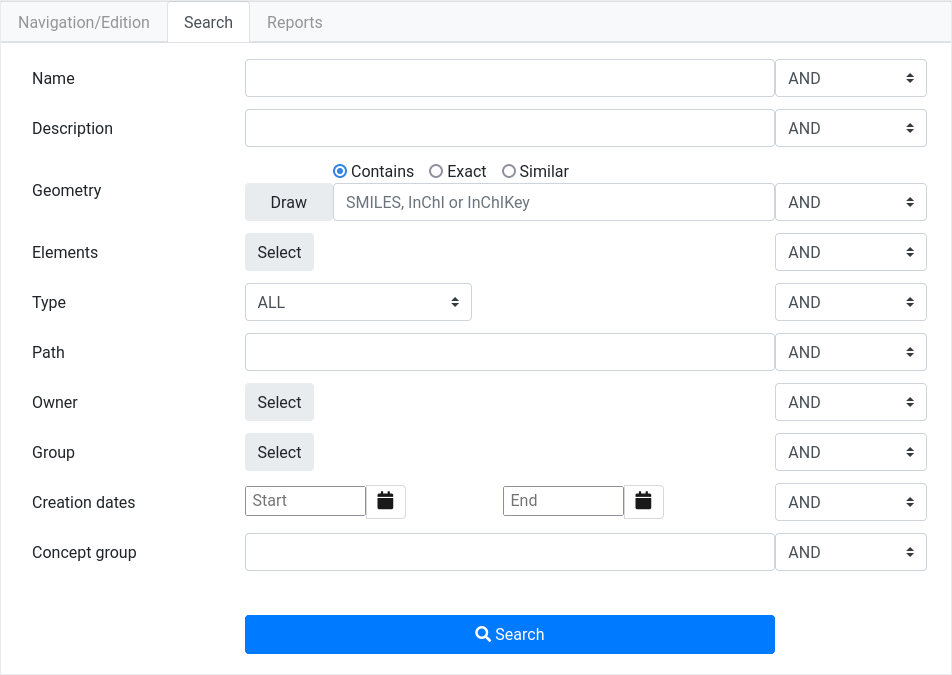
Search mode
The Search tab will display a form where the user can query all
uploaded projects/calculations. Multiple filters are available to
refine queries. Currently, it filters by administrative metadata and
molecular structure:
Name and description of the element, partial text matching search.
Type of the element (calculation software type).
Path, starting path to search for, see more at path definition section.
User who created the element and group of users that element belongs to.
Access permissions assigned to element.
Creation and modification dates.
Current element state. |
 |
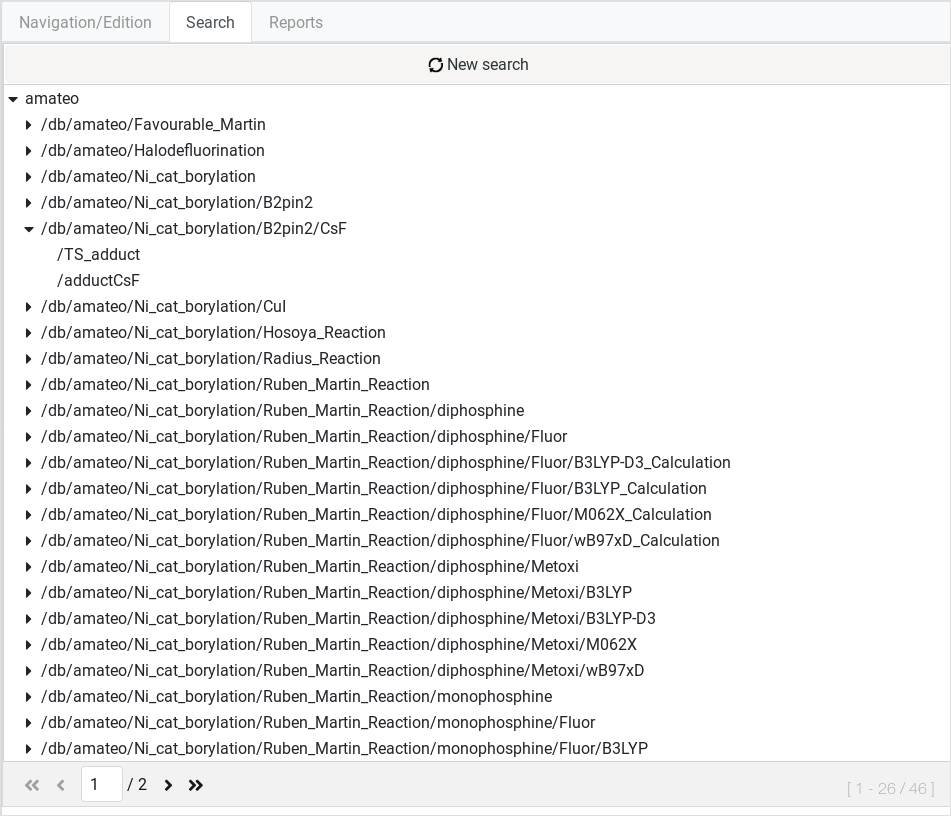
After setting the search parameters you whish, click the Search button to perform a query. A list of results will appear.
We can click on any listed element to display its particular Item
actions and details. To perform a new query, the Reset search button
will clean the form and will bring you back to Search form. |
 |
Report mode
This tab activates advanced operatives to process sets of calculations.
This mode is fully reported in the Generating reports page. Please
check it. |
Item Actions frame
This part of Create is in charge of displaying all actions related to a
selected element in the Navigation/Search frames. Projects do not have
any specific action associated. By selecting a project, it will display
the initial news page.
Calculations have the following actions, which are accessible in the tabs of this frame:
- 3D structure
- View results
- Download
- RAW CML
|
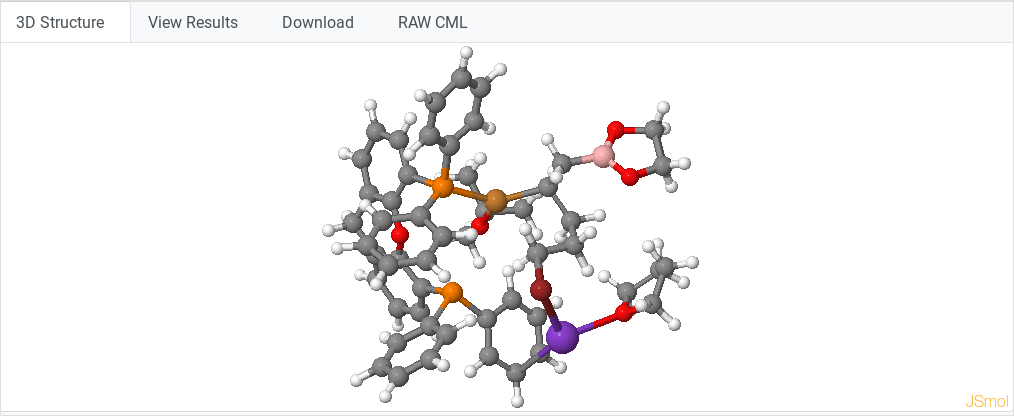
3D structure
This action displays the molecular structure or simulation cell. In the
case of geometry optimization runs, it will show the final geometry.
In the case of NEB calculations run with VASP, a special tab will allow visualizing all the points.
We use JSmol (Javascript version of Jmol) to display such structures
(and their cells in the case of periodic systems). Common JSmol
operations:
- Hold left click + drag = Rotate molecule
- Central button scroll = Zoom in / out
- Hold Shift + double click + drag = Translate molecule
- Right click = Display JSmol options menu
|
 |
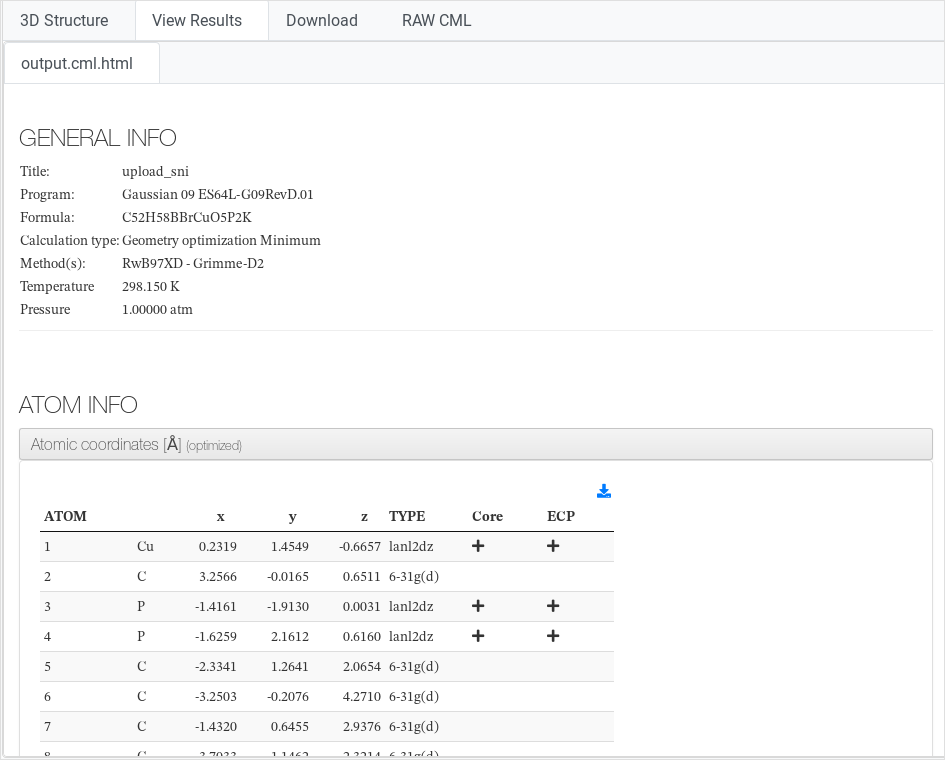
View Results
Some results of the output file uploaded to ioChem-BD are translated
into CML (Chemical Markup Language), an XML language oriented to
chemistry. Such markup language allows easy further conversion into any
existing format, such as an HTML5 report, a PDF file, or a JSON file.
Selected data is visualized, normally organized by content:
- General info: Contains calculation administrative and
descriptive metadata such is: user name, calculation type, methods
used, ...
- Settings: (VASP only), most relevant INCAR settings.
- Atom info: Atom type, coordinates and basis used. Eventually cell parameters, lattice vectors and atom valence.
- Molecular info: Implicit Solvation parameters, charge and multiplicity
- Job: Its content varies depending on the quantum
chemistry package used to generate the calculation (ADF software
generates different output fields than VASP, for example). For
calculations with multiple jobs this section will appear more than once.
There is an exhaustive description on which fields are captured and how
they are visualized. Please refer to Conversion template reference |
 |
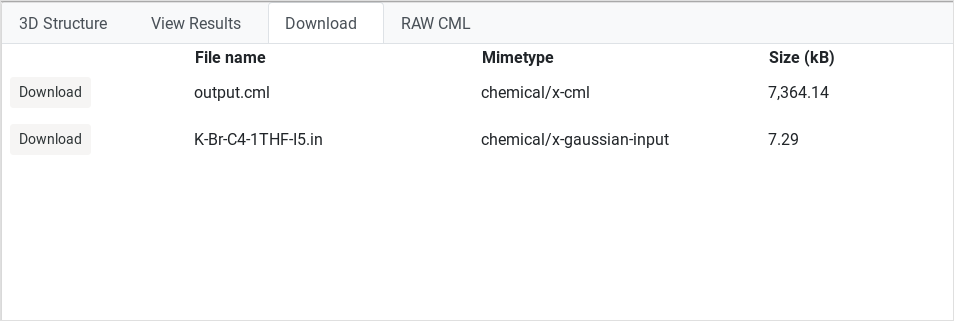
Download
This action allows downloading calculation files to your local filesystem. |
 |
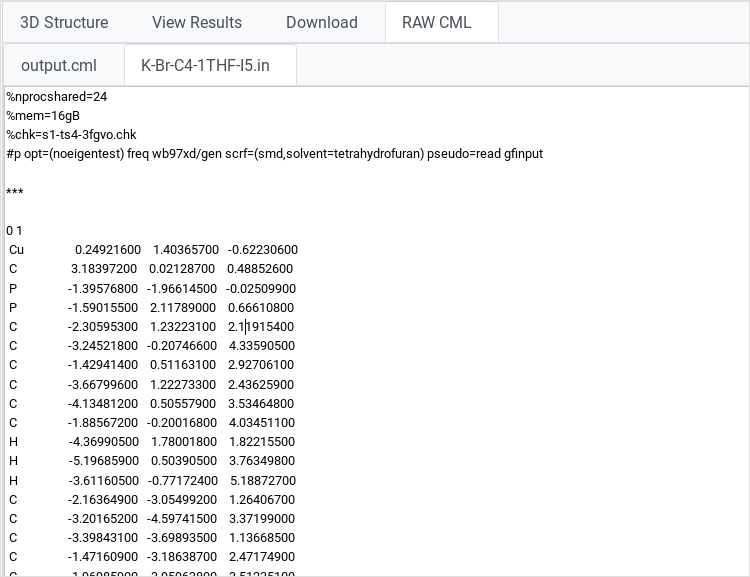
RAW view
This action displays calculation files content inside a text area. On
large files, it will start to download the file instead of displaying
it. |
 |
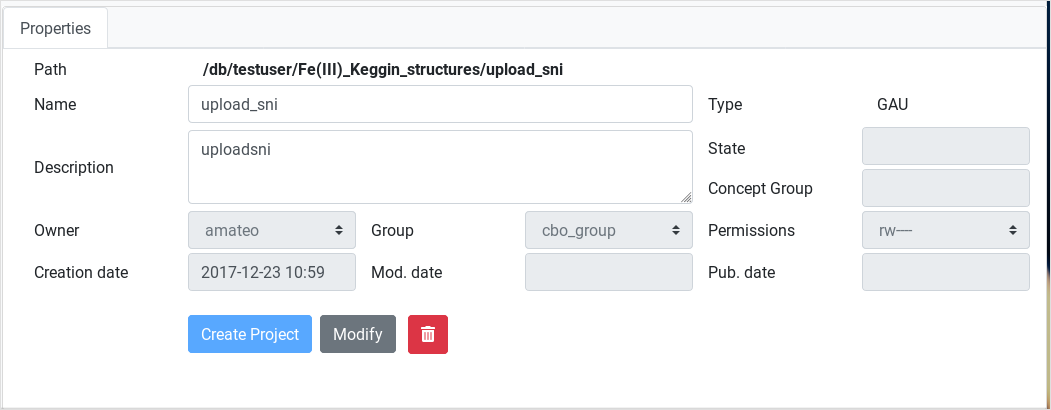
Item Details frame
Any project or calculation selected in our workspace will immediately
refresh this form, displaying its administrative metadata. Some fields
are modifiable f. ex. name, description, owner group, assigned
permissions. Other fields are fixed like owner user or creation date.
In the lower area of this frame three buttons provide important actions:
- Create project : Generates a project in the current path using the values in the form
- Modify : Replaces stored values for the selected element with values in the form
- Delete : Deletes selected element.
|
 |
Upload bar
On the bottom area of this frame there is an empty space. It is left
blank intentionally to fit the upload bar. This bar will only appear
while uploading calculations via the web interface, showing the
progress of the uploading step.. |
 |
ioChem-BD know More
|
|
Publish data to Browse know More
|
How to cite know More
|
Generating reports know More
|
|
|
|













